The Rpf2-Rrs1 complex in ribosome biogenesis
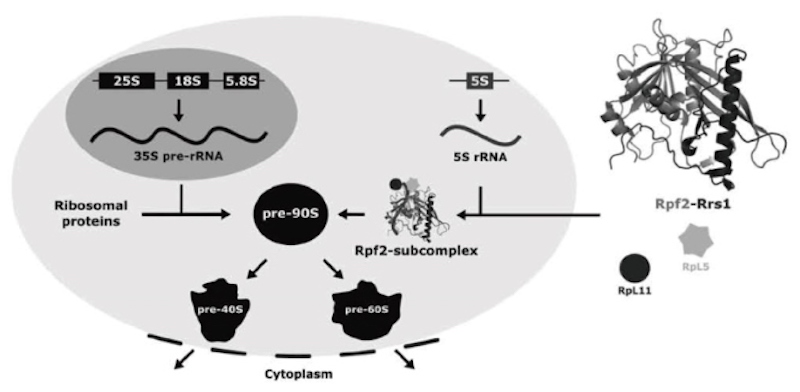
The ribosomes are highly conserved nucleoprotein particles which mediate the translation of genome information to proteins in all cells. Eukaryotic ribosomes are comprised of four different ribosomal RNAs (25S, 18S, 5.8S and 5S rRNA) and approximately 80 ribosomal proteins. The biogenesis of this large nucleoprotein particle is started at nucleus by transcription of three rRNAs (25S, 18S, 5.8S 25S) and then the ribosome precursor pre-90S is formed with ribosomal proteins (figure left).
Subsequently, maturation processes lead to formation of two independent complexes, pre-40S and pre-60S particles from matured pre-90S. Such two particles are transported to cytoplasm, and finally maturated to 40S and 60S with processing steps, respectively. Rpf2 and Rrs1 act as ribosome assembly factors which recruit RpL5, RpL11 and 5S rRNA into pre-90S by formation of Rpf2-subcomplex (figure middle) and function as checkpoint of maturation of pre-90S and pre-60S.
We determined the structure of Rpf2-Rrs1 core complex (fig. right), performed the mutants analysis and modeled a structure of pre-90S with Rpf2-subcomplex. Based on these studies and previous reports, we proposed a model for ribosomal component recruitment to pre-90.
Laboratory of NMR structural biology
Proteins modulate their activities by changing their structures. Many of the multi-domain proteins are known to undergo drastic structural changes to regulate their activities. Here we have demonstrated a strategy exploiting NMR spectroscopy with the paramagnetic lanthanide probe to visualize drastic conformational changes of multi-domain proteins. The paramagnetic lanthanide ion fixed on the protein induces several paramagnetic effects to the observed nuclei, including pseudocontact shift (PCS) that provides long-range (~40 Å) distance and angular information. By the use of PCSs, we succeeded to visualize conformational changes of MurD, a 47 kDa-bacterial enzyme consisting of three domains, coupled with the binding to the ligands and the inhibitors1. Since MurD is involved in the synthesis of bacterial cell wall, our results are expected to promote the development of new antibiotics.
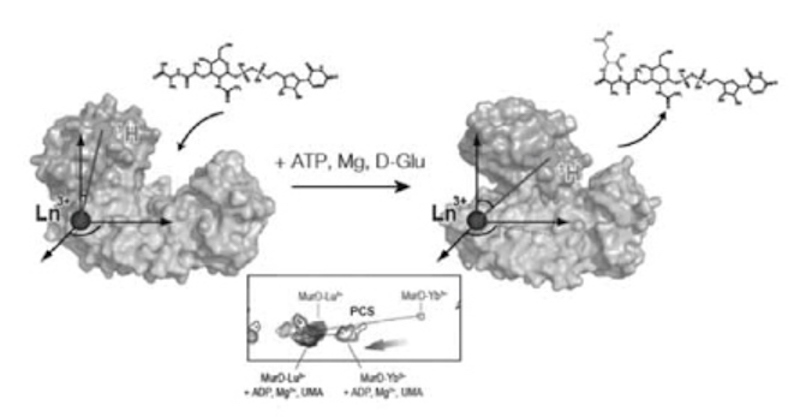
1. Ligand-driven conformational changes of MurD visualized by paramagnetic NMR.
Saio T, Ogura K, Kumeta H, Kobashigawa Y, Shimizu K, Yokochi M, Kodama K, Yamaguchi H, Tsujishita H, Inagaki F.
Sci Rep. 2015, 5, 16685