Laboratory of Photobiology
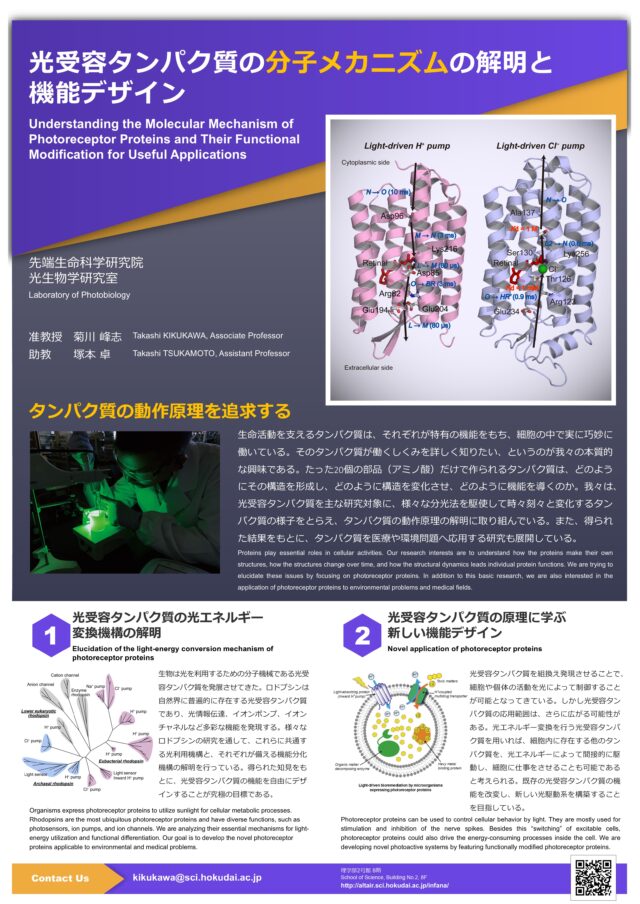
Elucidation of molecular mechanisms of photoactive proteins and their useful applications
- Research Theme
- Elucidation of the molecular mechanism of photoreceptor proteins / Functional modification of photoreceptor proteins for useful applications
- Research Keywords
Photoactive proteins, photobiology, rhodopsin, optogenetics, directed molecular evolution, membrane protein, ion pump, membrane transport, transporter
Staff
Overview of Research and Education
Organisms express photoactive proteins to utilize sunlight for cellular metabolic processes. Rhodopsins are the most ubiquitous photoactive proteins and have diverse functions, such as photosensors, ion pumps, and ion channels. We are analyzing their essential mechanisms for light-energy utilization and functional differentiation. Our goal is to develop the novel photoactive proteins applicable to environmental and medical problems.
Laboratory of Photobiology (pdf)
Charge
- Charge (US):
School of Science, Biological Science course (Macromolecular Functions), Core Laboratories - Charge (GS):
Graduate School of Life Science, Division of Soft Matter, Biomolecular Soft Matter
Contact
- Address
- 〒060-0810
N10, W8, Kita-ku, Sapporo, Hokkaido - Phone
- 011-706-3435
- kikukawa*sci.hokudai.ac.jp (Please replace * with @ when sending e-mail.)
Representative Publications
- Hamada, C., Murabe, K., Tsukamoto, T., and Kikukawa, T., Direct detection of the chloride release and uptake reactions of Natronomonas pharaonis halorhodopsin. J. Biol. Chem. 300, 107712 (2024)
- Sato, Y., Hashimoto, T., Kato, K., Okamura, A., Hasegawa, K., Shinone, T., Tanaka, Y., Tanaka, Y., Tsukazaki, T., Tsukamoto, T., Demura, M., Yao, M., and Kikukawa, T., Multistep conformational changes leading to the gate opening of light-driven sodium pump rhodopsin. J Biol Chem 299, 105393 (2023)
- Ohki, Y., Shinone, T., Inoko, S., Sudo, M., Demura, M., Kikukawa, T., Tsukamoto, T., The preferential transport of NO3- by full-length Guillardia theta anion channelrhodopsin 1 is enhanced by its extended cytoplasmic domain, J. Biol. Chem. 299, 105305 (2023)
- Doi, Y., Watanabe, J., Nii, R., Tsukamoto, T., Demura, M., Sudo, Y., Kikukawa, T., Mutations conferring SO42− pumping ability on the cyanobacterial anion pump rhodopsin and the resultant unique features of the mutant, Sci. Rep. 12, 16422 (2022)
- Sasaki, S., Tamogami, J., Nishiya, K., Demura, M., Kikukawa, T., Replaceability of Schiff base proton donors in light-driven proton pump rhodopsins, J. Biol. Chem. 297, 101013 (2021)
- Kato, T., Tsukamoto, T., Demura, M., Kikukawa, T., Real-time identification of two substrate-binding intermediates for the light-driven sodium pump rhodopsin, J. Biol. Chem. 296, 100792 (2021)
- Kikuchi, C., Kurane, H., Watanabe, T., Demura, M., Kikukawa, T., Tsukamoto, T., Preference of Proteomonas sulcata anion channelrhodopsin for NO3− revealed using a pH electrode method, Sci. Rep. 11, 7908 (2021)
- Murabe, K., Tsukamoto, T., Aizawa, T., Demura, M., Kikukawa, T., Direct Detection of the Substrate Uptake and Release Reactions of the Light-Driven Sodium-Pump Rhodopsin, J. Am. Chem. Soc. 142: 16023-16030 (2020)
- Iizuka, A., Kajimoto, K., Fujisawa, T., Tsukamoto, T., Aizawa, T., Kamo, N., Jung, KH, Unno, M., Demura, M., Kikukawa, T., Functional importance of the oligomer formation of the cyanobacterial H+ pump Gloeobacter rhodopsin, Sci. Rep. 9: 10711 (2019)
- Tsukamoto, T., Kikuchi, C., Suzuki, H., Aizawa, T., Kikukawa, T., Demura, M., Implications for the impairment of the rapid channel closing of Proteomonas sulcata anion channelrhodopsin 1 at high Cl- concentrations, Sci. Rep., 8: 13445 (2018)